About us
Our computational group is located in the Donnelly Centre at University of Toronto.
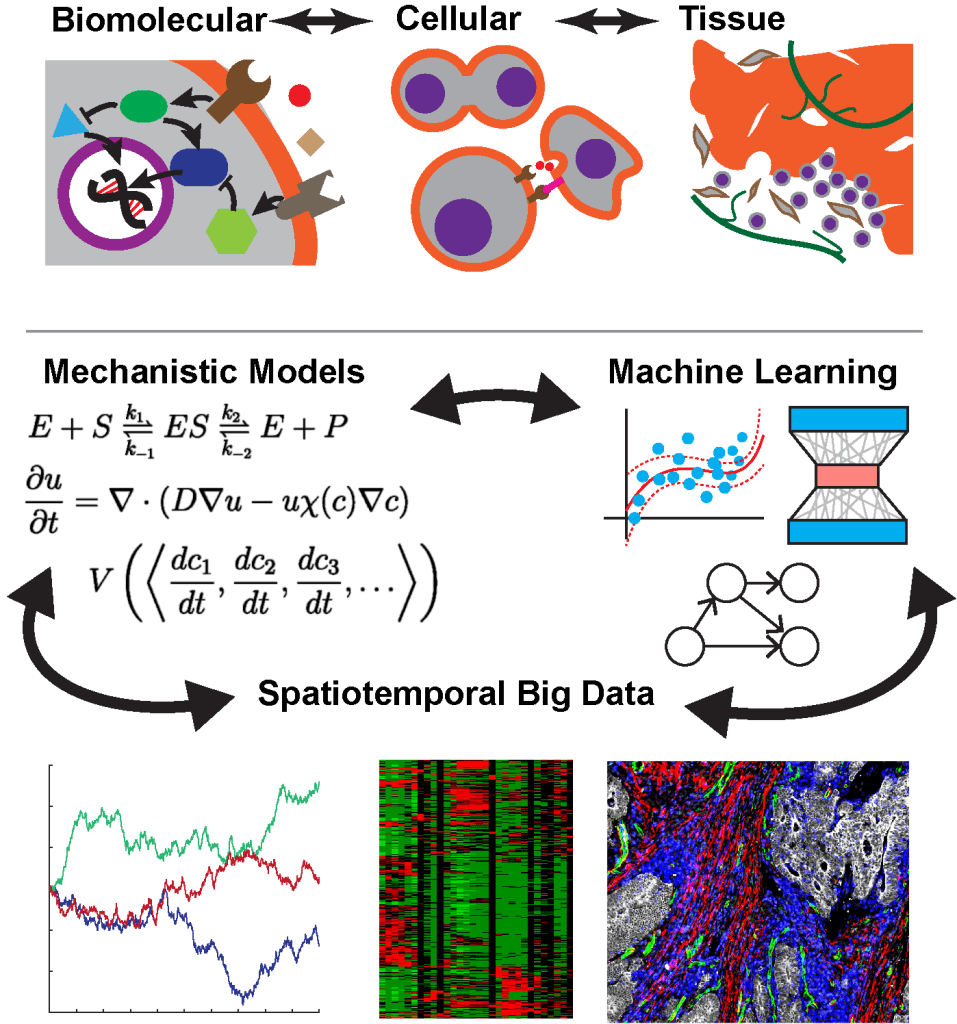
We are broadly interested in mathematically understanding the multi-scale networks underlying biological systems (e.g. protein signaling, cell states, tissue organization), seeking the accuracy and precision that would be needed to reliably treat heterogeneous diseases such as cancer. To this end, we emphasize both empirically analyzing high-dimensional, high-throughput biological data (e.g. single-cell multiplex imaging or spatial transcriptomics), as well as mathematically analyzing mechanistic models of biological processes, using whatever tools are appropriate case-by-case.
In the past, data analysis techniques have included supervised learning, manifold learning, spatial statistics, and probabilistic graphical models. Past mathematical modeling techniques have included dynamical systems theory, differential and algebraic geometry, and chemical reaction network theory.